Exploring the structural and functional dynamics of trimeric and tetrameric states of influenza encoded PB1-F2 viroporin through molecular dynamics simulations
Exploring the structural and functional dynamics of trimeric and tetrameric states of influenza encoded PB1-F2 viroporin through molecular dynamics simulations.
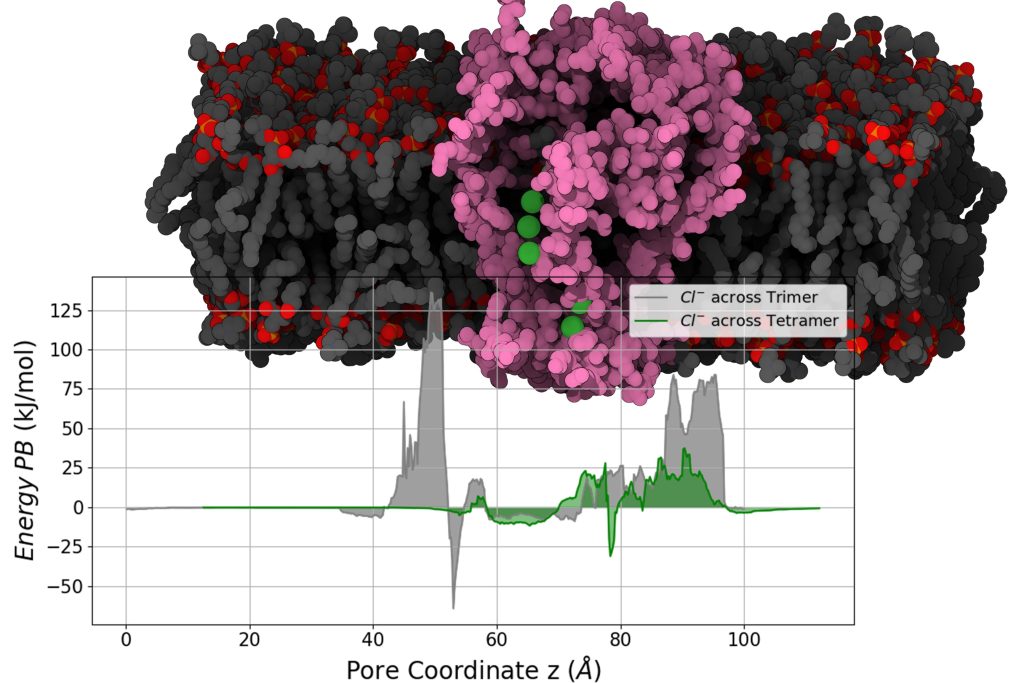
Viruses act by hijacking the host cellular functions to replicate and evade the immune system. They utilise specialised small proteins that contain a cluster of basic amino acid residues along with amphipathic-α-helices, widely known as viroporins. However, the elusive nature of viroporins complicates the efforts to comprehend their mechanisms. One such type of protein is ‘PB1-F2’ encoded by the influenza virus, recognized for its pro-apoptotic properties, despite several scientific investigations their exact mode of action at the molecular level in an apoptotic pathway is still unclear. Our research investigates whether PB1-F2 can form pore-like structures in cell membranes, which could allow ions and other molecules to pass through, potentially influencing viral infections. Using molecular modeling and molecular dynamics simulations, we analysed different structural forms of PB1-F2, more specifically its trimeric and tetrameric states, in a host membrane environment. We found that the tetrameric form appears to be more stable and functionally suited for ion transport, a key feature of viroporins.
By understanding how PB1-F2 interacts with membranes and contributes to cell death, our findings provide crucial insights into its role in influenza infections. This paper could help develop new antiviral strategies targeting PB1-F2, potentially weakening the virus and improving treatments for flu-related illnesses.
Authors: Sehrish Jamal, Syed Tarique Moin, Shozeb Haider
